Features
General Features

Python Scripting
|

Program in C++
|

command line interface for batch processing
|
graphical user interface (DNG)
|
License
OpenStructure is released under the
LGPL Version 3 license.
|
|
|
Macromolecules
|
Molecular structures are a central aspect of OpenStructure and a rich understanding of macromolecular structure datasets with access to atoms, residues, chains, bonds and torsions is offered.
|
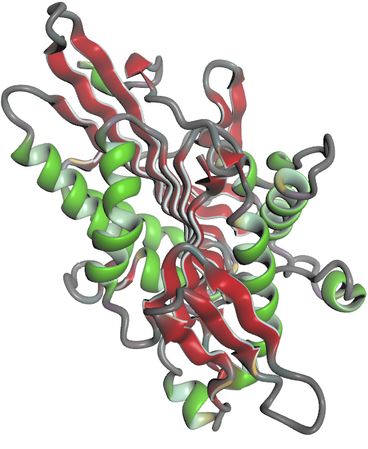
|
|
|
Scoring Against Reference |
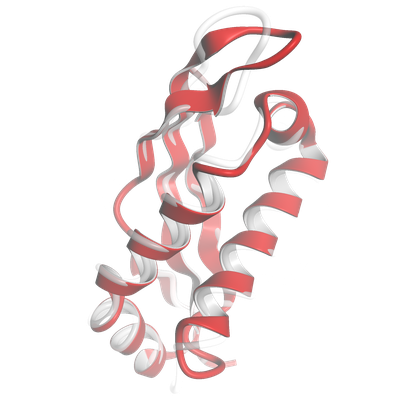
|
- Comparing a model against a reference with a wide variety of scores.
- Oligomeric scores for macromolecular complexes: lDDT, QS-score, DockQ, GDT, RMSD, and more.
- Ligand scores for small molecules: lDDT-PLI, RMSD.
- Programmatic API (Scorer for
oligomeric macromolecules and
LigandScorer for small molecules).
- Command line interface with dedicated actions to compare
two macromolecular structures or two
structrures with ligands.
|
Density Maps and 2D Images
Sequences And Alignments
- Tight integration with molecular structures: Map back and forth between structures and alignments.
- Calculate conservation scores from alignments.
- Convenient methods to manipulate alignments: Cutting, shifting, and removing of columns.
- file formats.
|
Real-time Rendering
- Rendering of molecular structures, surfaces, density maps and geometric Objects.
- Rendering parameters such as colring can be adjusted programmatically and in a convenient user interface.
- Experimental export to POVray.
|
|
Features
|